Deargen was selected as Top Performers of the NCI-CPTAC DREAM Challenge
Deargen was selected as a Top Performer of the NCI-CPTAC DREAM Proteogenomics Computational Challenges that is international Challenges for cancer proteome prediction. Details of the challenge will be published in the Nature Method.

The Dream Challenges is a collective intelligence research group that researchers from around the world are trying to solve difficult problems in biomedical fields through competition and collaboration and it has held a challenge since 2007. Research results from the challenge are being published in the highest authoritative journals such as Nature, Cell, and Science. World-renowned research institutions such as the National Institutes of Health (NIH), the Sanger Institute (in the UK), and the IBM Institute, etc. host and supervise the this challenge.
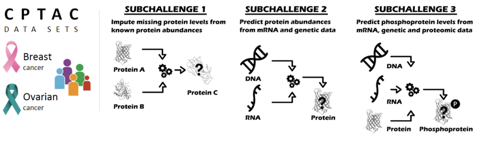
In this challenge, the NCI-CPTAC (The National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium) provided data and presented three questions that predict the degree of protein activation in ovarian and breast cancer patients. Among them, we were selected as a Top Performer for the second question.
- Can one impute missing values in proteomics data given observed proteins?
- Can one predict abundance of any given protein from mRNA and genetic data?
- Can one predict the phosphoproteomic data, using proteomic, mRNA and genetic data?
We has developed an algorithm that can predict protein expression levels in breast and ovarian cancer patients by using the Ensemble Technique of Machine Learning. To solve problems such as overfitting, etc. that can occur during a model learning with a small number of data and to improve the performance of the model, learning was progressed by groups after grouping proteins with similar amounts of the expression level of protein coding genes. Also, it enables accurate prediction of protein expression level based on various characteristics by making the information obtained based on machine learning as well as the genomic information of patients be used for learning.
Researcher Lee Bora who has conducted research said, “We have shown that machine learning can be effectively applied to biomedical fields where it is difficult to collect data. I would like to go one step further in patient-specific diagnosis with an accurate prediction of protein expression level in cancer patients.”
We consider it meaningful to make an outstanding achievement of being selected as a Top Performer in the competition with over 60 world-class research teams, including UCLA and Stanford, etc. We will continue to work with a variety of partners in the field of drug development and create artificial intelligence models that can impact the advance era of the precision medicine.
The results of this Dream Challenge will be published in the Nature Method, which can be available through the attached link later.