A new state of the art drug target interaction model, Molecule Transformer based Drug Target Interaction (MT-DTI) Model.
Deargen Presented a world-class drug-protein interaction prediction model, MT-DTI (Molecule Transformer Drug Target Interaction Model). This research is published in the Journal of Machine Learning Research (JMLR).

Introduction
Drug Target Interaction (DTI) plays a key role in the drug discovery process. This is because it selects drug candidates that regulate abnormal protein levels that cause disease from a large group of drug candidates. It cost a lot of money and take a lot of time because in-vitro DTI has been implemented traditionally. However, in-silico-based DTI models have emerged in recent years, so attempts for effective drug discovery processes are being made.
In these attempts, various technologies based on machine learning and deep learning have been proposed, but it has been known that those technologies have limitations in predicting candidates because they do not properly reflect complex chemical structures and do not use extensive compound information. To solve these problems, in this article, we modeled complex chemical structures more effectively by applying the self-attention mechanism to the DTI model and improved the accuracy of the model by using PubChem’s database of approximately 97 million compounds for pre-learning.
Background

[Figure 1. The Drug target Interaction (DTI) task]
A DTI model takes a drug and a protein as an input pair and predicts a affinity score between the two inputs (Figure 1). Both a drug and protein come in the form of a sequence. A drug is a sequence called SMILES that contains atoms and binding information, and protein is a sequence called FASTA that consists of an array of amino acids. With this formalization, the DTI task can be generalized into a regression problem of the machine learning so that different approaches can be tried out. For example, the most recent DTI model before this paper is DeepDTA that is based on a 2D convolutional operation.
Motivation
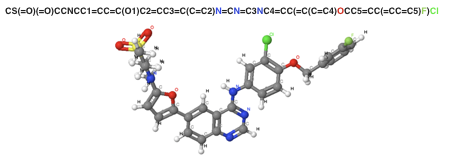
[Figure 2. SMILES Sequence notation and 3D structure in Lapatinib]
In general, a convolutional operation considers adjacent tokens in a sequence. However, there is no guarantee that useful information will always be in adjacent tokens. For example, according to Figure 2 that shows the three-dimensional structure and the sequence notation of a well-known anticancer drugs, the adjacent pair, Cl and F will be convoluted together while CI and N will never be due to the long distance in the sequence representation. However, the actual distance between CI and N is closer than the distance of Cl and F in the graphical representation. Therefore, this approach is limited to capture precise information when representing a molecule.
Besides, existing DTI models have structural problems that can’t have access to a large database of small molecules. Therefore, in this paper, we not only made all tokens be effectively considered by applying the self-attention mechanism to the model but also applied knowledge of compound synthesis patterns that were efficiently extracted from existing small molecules databases to the model.
Method

[Figure 3. Molecule Transformer]
In order to effectively model small molecules, we have proposed a Molecule Transformer (MT, Figure 3). It consists of a stack of several encoders based on self-attention. Pre-training was performed on PubChem’s approximately 97 million compounds in order for MT to train key knowledge such as syntax and semantics of small molecules. As a result, it was able to perform the grammar prediction task of the SMILE sequence with more than 97% accuracy.

[Figure 4. MT-DTI Model]
The fine-tuning on DTI datasets is performed by transferring this trained MT into the DTI model of Figure 4 as it is. This process of doing fine-tuning by putting a part of a model into a large model after pre-training is called transfer learning. This is very useful when you don’t have enough data related to what you want to do. This is because pre-trained models are not the data for the desired task, but important information from a vast amount of data similar to the data can be obtained.
Experiments

A comparative research on four performance measures about the two data was performed. Both Kiba and Davis are DTI data and consist of a score indicating the degree of binding that is each criterion for drugs and proteins. The value CI (concordance index) and r ^ 2_m are the success measures for regression, MSE represents the difference between the predicted value by the model and the actual value, and AUPR indicates the accuracy of predicting whether the binding is shown by replacing the regression with a binary problem.
The model presented in this paper is MT-DTI, and the model with w / o FT is the MT-DTI model that fine-tuning is omitted. The latest machine learning and deep learning-based models were used for the comparison. Research results showed that the MT-DTI had superior performance in all performance measures compared to the existing methods. Also, even when fine-tuning is omitted, it was similar or sometimes better than the existing methods. This suggests that the self-attention based molecule representation using pre-training effectively learns useful chemical structure that can be exploited by the interaction model.
Case Study

This performance comparison experiment showed that the proposed MT-DTI model can accurately predict binding scores. Not only that, we further tried to figure out whether this model could actually be used in drug development through a case study.
We performed a case study using actual FDA-approved drugs targeting a specific protein, epidermal growth factor receptor (EGFR). This protein is chosen because this is one of the famous genes related to many cancer types. We calculated the interaction scores between EGFR and the 1,794 selected molecules based on the Drugbank database. Surprisingly, a result showed us that all eight of the actual EGFR target drugs existed in the Drugbank were among the candidates. This not only proved the usefulness of the proposed model but also meant that 22 other proposed drug candidates need to be tested as potential anticancer drugs.
Conclusion
The proposed MT-DTI model effectively introduces the latest deep learning technology by focusing on the limitations of the existing DTI models and showed the best performance in two standardized DTI data. It also showed that the proposed model through a real case study could be used effectively in the process of drug discovery. However, the self-attention was only applied to the side of the drug that is one of the inputs of the DTI. But, if it is extended to proteins, a better model can be expected.
To do this, a large number of protein datasets are needed to be secured. Also, self-attention can be only applied to short sequences, so if a new model that can handle long sequences effectively is developed, a better DTI model can be expected.
Test Our MT-DTI
MT-DTI model can be used through the button below. It’s easy to check the DTI score of your own data.